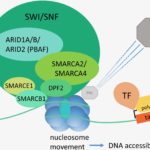
ARID1A has the highest mutation rate across all components in the SWI/SNF complex. ARID1A mutations occur in many types of human cancers and are notably one of the most frequently mutated genes in cancers. For instance, ovarian clear cell carcinoma has a 50% mutation rate [2] and colorectal adenocarcinomas have an 11% mutation rate [1]. SWI/SNF complex mutations are found in 20% of all types of human cancer [3]. Compared to the SWI/SNF complex, the polycomb repressive complex 2 (PRC2) has been studied well within the context of cancer biology. There is speculation that there is an antagonistic relationship between Polycomb group Proteins and the SWI/SNF complex [6]. Consequently, there is an interest in understanding whether the interaction between the PRC2 and SWI/SNF complex plays a functional role in shaping cancer immune phenotype and T cell immunity (figure 1) [4].

The catalytic subunit of the Enhancer of Zeste 2 PRC2 (EZH2) functions as a methyltransferase. EZH2 mediates gene repression and plays an oncogenic role in a variety of cancers. It has been shown that EZH2 represses Th1-type chemokine expression and alters effector T cell tumor trafficking [5]. A recent study on ARID1A epigenetic driver mutation’s role in shaping immune phenotype and immunotherapy, uncovered if there was an interaction between ARID1A and EZH2 that affects T cell immunity and if ARID1A mutations functionally altered the interaction between the ARID1A and EZH2 interaction in tumors [4].
ARID1A and Patient Survival
ARID1A gene status affects patient survival. There was a noticeable outcome when the analysis was run over several types of cancers and the ARID1A gene status. The data suggest that ARID1A gene status can affect the patient outcome in multiple types of cancer. For instance, patients with ovarian cancer and ARID1A-mutated tumors had poor overall survival compared to patients whose tumors had WT ARID1A (figure 2A); and ARID1A mutations were also associated with poor survival in patients with hepatocellular carcinoma (figure 2C) [4].
There are found correlations between ARID1A gene status and IFN signaling gene signature, T cell tumor infiltration, patient survival, and clinical responses to checkpoint blockade in several types of cancer regardless of tumor mutation load. Recent findings support that driver mutations can directly affect tumor T cell immunity and immunotherapy. Cancer epigenetic driver mutations such as ARID1A mutations, shape tumor immune phenotype, and drive cancer immune evasion in many types of cancer [4].

ARID1A Shapes Cancer Immune Phenotype and Immunotherapy
Regardless of the tumor mutation load, ARID1A mutations were associated with poor clinical benefit in patients who received immunotherapy. The nature of immunosuppressive mechanisms, tumor epigenetics, and tumor metabolism all have a role in determining cancer-immune phenotype and immunotherapy response. Current findings suggest ARID1A gene status has the potential to affect spontaneous and immune checkpoint blockade-induced T cell immunity.
Current findings support ARID1A gene status affects cancer immune evasion. ARID1A controls IFN signaling gene chromatin accessibility, which regulates tumor cell response to spontaneous and therapy mediated immune stimulation [4]. Therefore, targeting SWI/SNF complex mutation-associated pathways combined with immunotherapy may be a new approach to treating patients with SWI/SNF cancers. There is a need for additional investigation of the immunological role of each epigenetic component mutation in different types of cancer.
References
- Giannakis, M., Mu, X. J., Shukla, S. A., Qian, Z. R., Cohen, O., Nishihara, R., … Garraway, L. A. (2016). Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Reports, 15(4), 857–865. https://doi.org/10.1016/j.celrep.2016.03.075.
- Jones, S., Wang, T., Shih, I., Mao, T., Nakayama, K., Roden, R., . . . Papadopoulos, N. (2010). Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science (New York, N.Y.), 330(6001), 228-231. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3076894/
- Kadoch, C., Hargreaves, D., Hodges, C. et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy.Nat Genet 45, 592–601 (2013). https://doi.org/10.1038/ng.2628.
- Li, J., Wang, W., Zhang, Y., Cieślik, M., Guo, J., Tan, M., … Zou, W. (2020). Epigenetic driver mutations in ARID1A shape cancer immune phenotype and immunotherapy. The Journal of Clinical Investigation, 130(5). https://doi.org/10.1172/JCI134402.
- Peng, D., Kryczek, I., Nagarsheth, N. et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 527, 249–253 (2015). https://doi.org/10.1038/nature15520.
- Wilson, B. G., Wang, X., Shen, X., McKenna, E. S., Lemieux, M. E., Cho, Y.-J., … Roberts, C. W. M. (2010). Epigenetic Antagonism between Polycomb and SWI/SNF Complexes during Oncogenic Transformation. Cancer Cell, 18(4), 316–328. https://doi.org/10.1016/j.ccr.2010.09.006.